Zohre Eskandari Alughare, Eduardo Antúnez de Mayolo, and Christopher Engelhard are working on this project.
Protein structural dynamics
The structural dynamic properties of proteins often play a crucial role in their functionality. Our main interest is in a) studying how – often thermodynamically driven – structural fluctuations inherent to proteins can influence their function and b) observing intermediate states in complex structural transitions by observation of single proteins over long time periods. To this end, we employ fluorescence spectroscopy in a confocal microscope, both at room temperature and in a cryogenic setup that allows us to observe biomolecules at any temperature, as well as create rapid temperature shifts, freezing and thawing dynamic fluctuations at will. This has, for example, allowed us to observe structural fluctuations of the small redox protein Cu-Azurin that directly influence its reaction parameters [1].
Thermal snapshots
One difficulty in observing structural fluctuations in proteins using fluorescence is that the time available to study a single biomolecule is limited by the excitation
laser bleaching the fluorophore. The experiment is therefore hampered by conflicting needs: The time available can be extended by using low temperatures and low laser power, but fluctuations require ambient temperatures, and a high time resolution requires high laser powers.
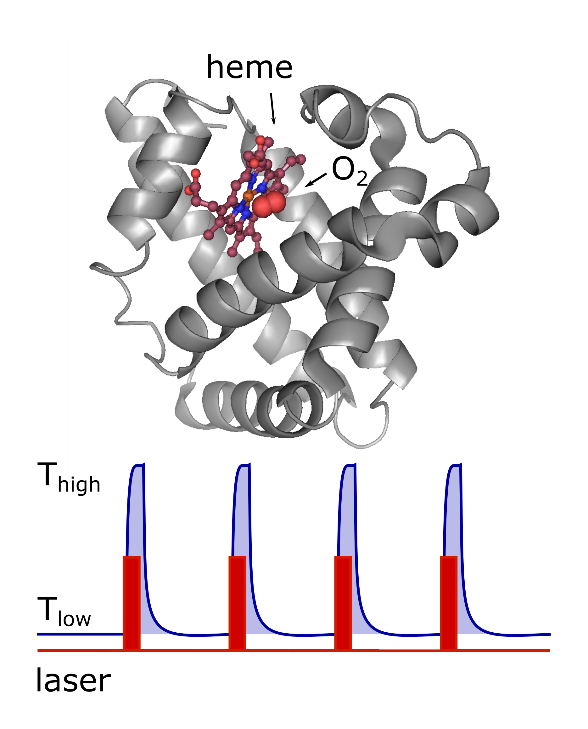
To work around this problem, this group has previously developed the thermal snapshot technique [2]. By using IR laser heating of a cryogenically cooled sample, we achieve the extreme temperature gradients necessary to essentially instantly stop and restart (internal) protein movements. This decomposes a dynamical process into a series of small movement steps interspersed with static (frozen) pauses. Measuring during those static phases, allows us to use low illumination at low temperatures, while the entire trajectory can be reconstructed from a series of such snapshots akin to a movie.
Additionally, precise and rapid temperature control enables us to probe highly temperature-dependent processes such as the temperature-induced unfolding of cold-shock proteins.
Myoglobin
One example of the processes we’re interested in is the binding of carbon monoxide to Myoglobin. Myoglobin as an oxygen storage protein plays a crucial role in living cells. There are many studies that investigate the mechanism and reversible kinetics of ligands binding to the heme cofactor. One striking aspect of these studies is that they seem to suggest that the rates of bond formation to these ligands, particularly CO, can vary by orders of magnitude [3]. Here, single-molecule spectroscopy is a powerful tool to visualize and analyze interactions and dynamics of individual molecules, while our cryogenic setup means that we can observe purely the rate of bond formation in the frozen sample while still allowing the protein to explore its conformational state by intermittently heating it to the liquid phase.
References
[1] Biswajit Pradhan, Christopher Engelhard, Sebastiaan Van Mulken, Xueyan Miao, Gerard W. Canters and Michel Orrit, “Single electron transfer events and dynamical heterogeneity in the small protein azurin from Pseudomonas aeruginosa”, Chem. Sci. (2020), 1.
[2] Rob Zondervan, Florian Kulzer, Harmen van der Meer, Jos A. J. M. Disselhorst, and Michel Orrit, “Laser-Driven Microsecond Temperature Cycles Analyzed by Fluorescence Polarization Microscopy”, Biophys. J. (2006), 90, 2958–2969.
[3] R. H. Austin, K. W. Beeson, L. Eisenstein, H. Frauenfelder and C. Gunsalus , “Dynamics of ligand binding to myoglobin”, Biochemistry (1975), 14, 5355-5373.


